1. Motivation
In the previous post, I described the procedures for combining molecular information from both EESI-TOF and Orbitrap measurements. In our offline system, EESI-TOF can be regarded as semi-quantitative and more stable for long-term monitoring, making it ideal for tracking consistent ion trends. One key purpose of Orbitrap data is to guide the identification and fitting of TOF-detected ions, using its ultra-high resolution and accurate mass information.
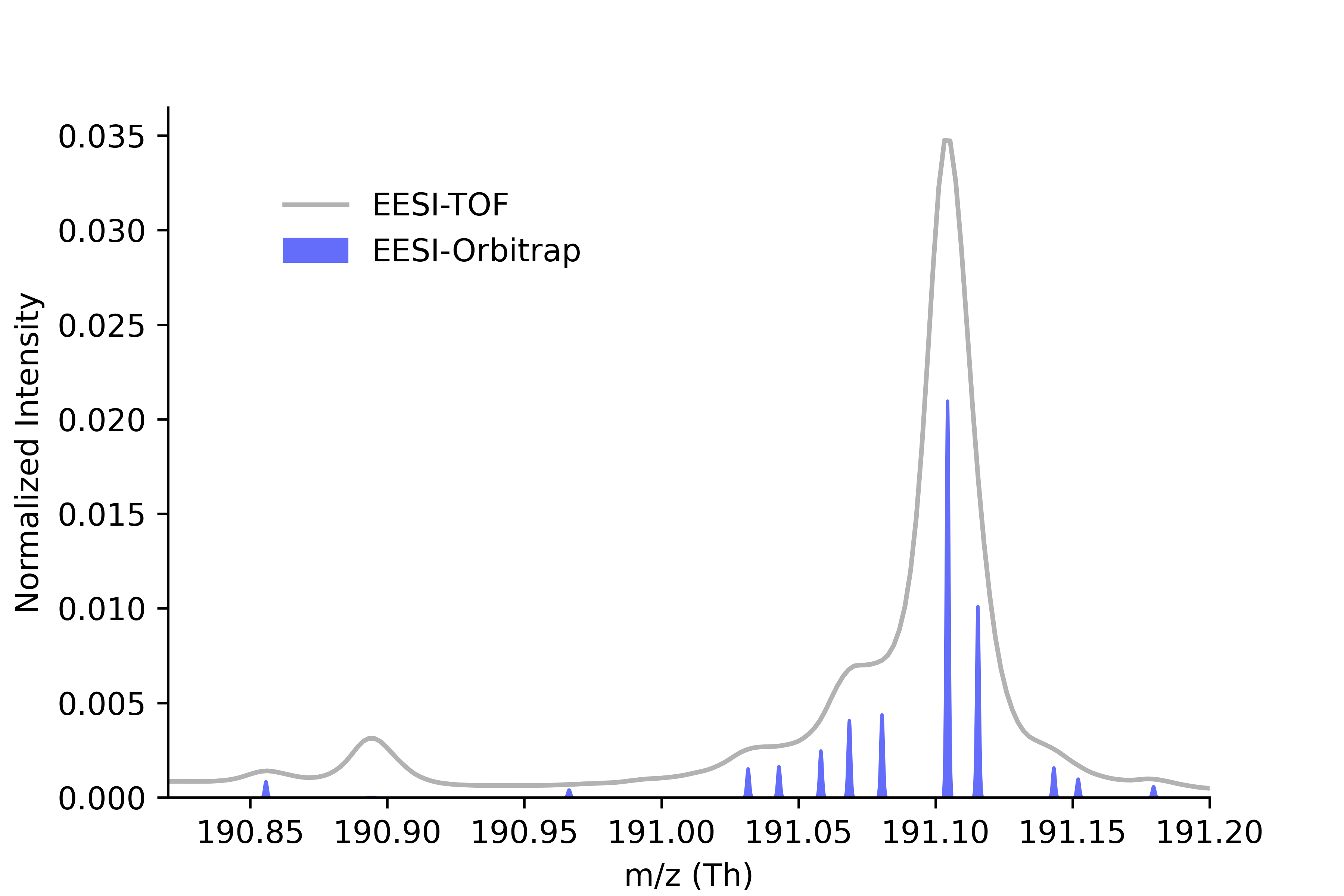
However, in some cases, certain peaks are clearly detected by TOF but are either missing in Orbitrap or filtered out during the Orbitrap peak filtering and clustering process. These are often real chemical features, especially in complex mixtures, that may be weak or distorted in Orbitrap but still prominent in TOF.
To address this, I implemented a strategy to embed missing TOF peaks into the final clustered peak list, ensuring a more complete molecular representation, especially important in non-targeted or mixture-rich analyses.
Below is an example where Orbitrap-resolved ions (blue bars) partially explain a broader TOF feature, while a peak near m/z 190.90 in which clearly visible in TOF trace were not captured by Orbitrap. it is likely a chemically real ion that should be integrated into the final peak list.
2. Code Strategy
The following logic checks for high-prominence TOF peaks that were not matched in Orbitrap clustering results, and adds them back into the final peak list as "Note": 'Missing Peak'.
The procedures can be described as follows with code also attached:
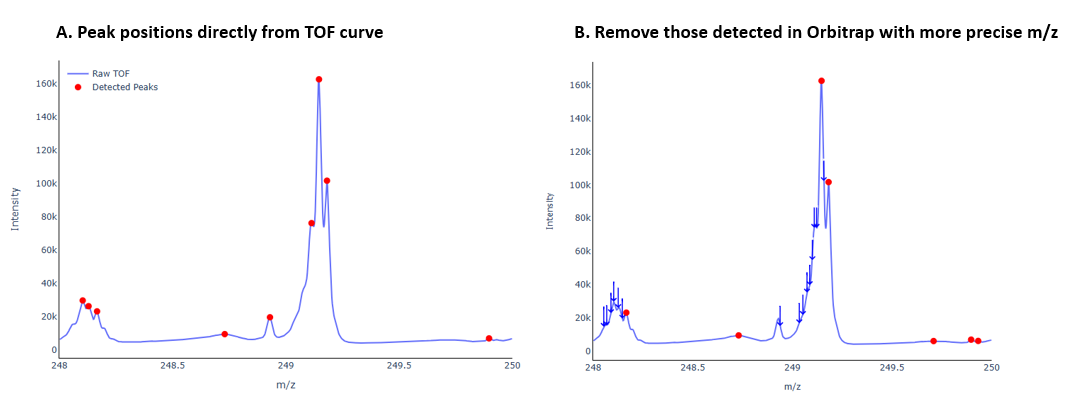
Peak detection from TOF
Usingfind_peaksfunctions from scipy library to search for the local maixma values. Consider that there are some peaks on the shoulder only larger than one side, to avoid overfitting, we fitltered to keep those with strong prominice on both left and rigth side.Tolerance Matching with Orbitrap Peaks
For each detected TOF peak apex, we check whether any Orbitrap-derived peak falls within a defined ppm tolerance window. If no match is found, the TOF peak is considered “missing in Orbitrap” and marked accordingly.
In the final step, we combine the Orbitrap-processed peak list with these unmatched TOF peaks, resulting in a more comprehensive ion profile.
3. Code
1 | def filter_asymmetric_peaks(peaks, props, y_data, left_thre=500, right_thre=500): |
Comments